id = 'stars'
site_name = 'Gila River Indian Community'
data_dir = 'gila-river'
download_key = 'gila-river-ndvi'
project_title = 'Gila River Vegetation'
start_year = 2001
end_year = 2022
event_year = 2012Vegetation Data Access
Accessing NDVI data on Tribal Subdivisions
For the vegetation health coding challenge, you used some sample data that we packaged up for you. In this lesson, we’’ll go through how we got that data. Give it a try, and then modify it to answer your own scientific question!
STEP 0: Set up
Import libraries
We’ll need some Python libraries to complete this workflow.
In the cell below, making sure to keep the packages in order, add packages for:
- Working with DataFrames
- Working with GeoDataFrames
- Making interactive plots of tabular and vector data
What are we using the rest of these packages for? See if you can figure it out as you complete the notebook.
import json
import os
import pathlib
from glob import glob
import earthpy.api.appeears as eaapp
import earthpy
import hvplot.xarray
import rioxarray as rxr
import xarray as xrSee our solution!
import json
import os
import pathlib
from glob import glob
import earthpy.api.appeears as eaapp
import earthpy
import geopandas as gpd
import hvplot.pandas
import hvplot.xarray
import pandas as pd
import rioxarray as rxr
import xarray as xrNext, we’ll set some parameters. You can use these to customize your workflow.
We have one more setup task. We’re not going to be able to load all our data directly from the web to Python this time. That means we need to set up a place for it.
- Replace ‘my-data-folder’ with a descriptive directory name.
- Run the cell to display your project directory.
- Can you find the directory, either in a terminal or through your operating system’s file browser/explorer/finder?
# Create a project directory in the system data folder
project = earthpy.project.Project(dirname='my-data-folder')
project.project_dirSee our solution!
# Create a project directory in the system data folder
project = earthpy.project.Project(dirname=data_dir)
project.project_dir
**Final Configuration Loaded:**
{}PosixPath('/home/runner/.local/share/earth-analytics/gila-river')STEP 1: Study area
You can use any boundary for your study. In the example, we used the “American Indian Tribal Subdivisions” available from the US Census.
We realize not everyone is cool with the term “American Indian”, especially when used by people who aren’t Tribal members. At the same time, others feel that it is important to reclaim “Indian”, and more importantly that’s the term a lot of Tribal people use every day. In this case, we’re stuck because of how this dataset is named in the Census boundary datasets…however, when sharing your work or publishing your own data, it’s important to include local communities in the area you are studying, about language choices but also throughout your project.
We’ll be downloading the boundaries using a web browser. The data source, data.gov, has placed restrictions on programmatic downloads (downloads using code). There are a number of ways to attempt to get around these restrictions, such as pretending to be a web browser by setting the User Agent parameter, but in this case we’ve found it’s just not worth the time and effort. Follow the steps below to download the data and put it somewhere that your Python code can find it.
STEP 1A: Open up the data catalog
Click this link: https://catalog.data.gov. You can also open your web browser and type it in if you prefer.
STEP 1B: Search
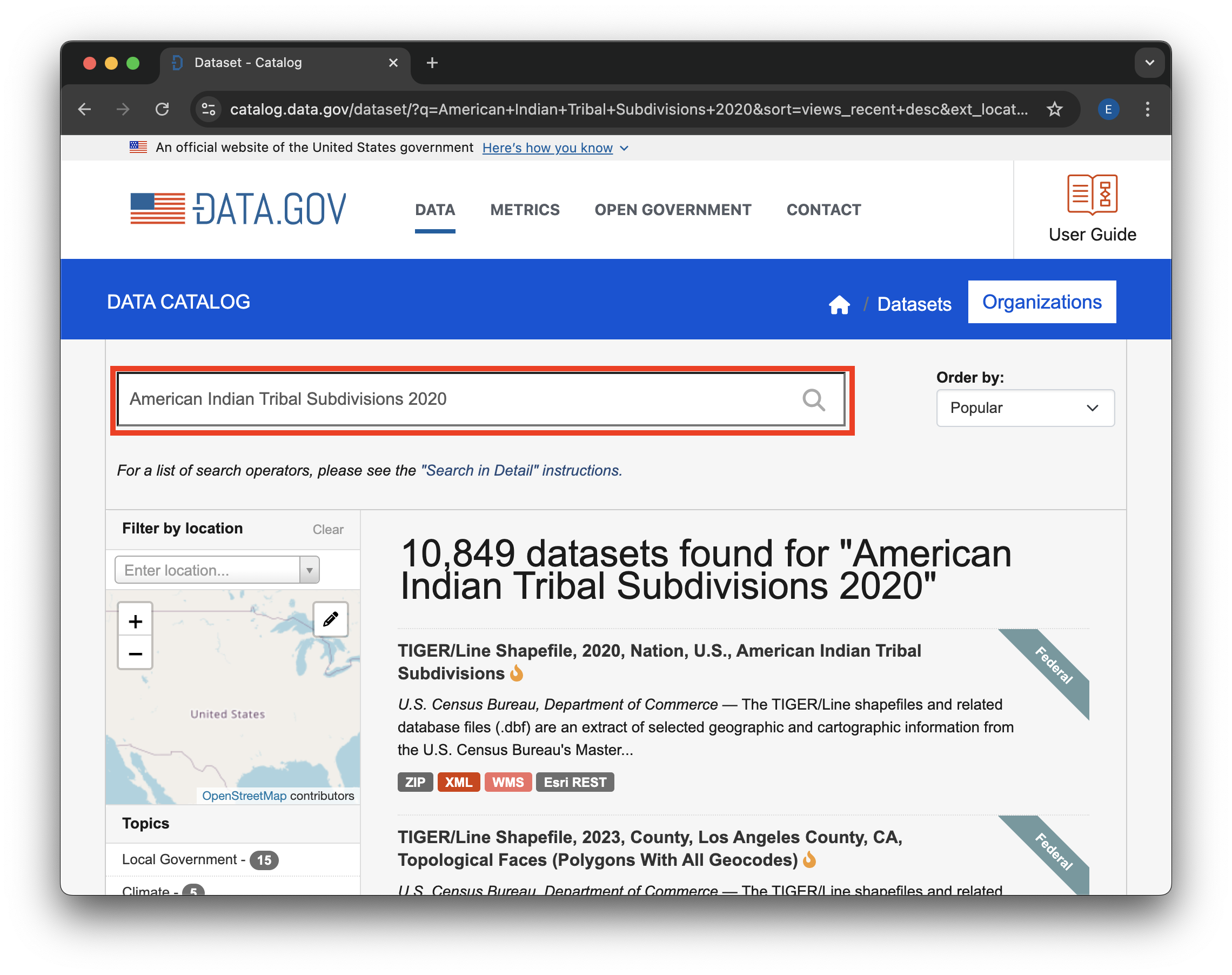
Search for the data you want. To get the American Indian Tribal Subdivisions census data, we recommend the search term “American Indian Tribal Subdivisions 2020”. These census files are updated regularly at the time of the census, so we want to make sure we have the most recent version. The last census was in 2020.
STEP 1C: Open the dataset page
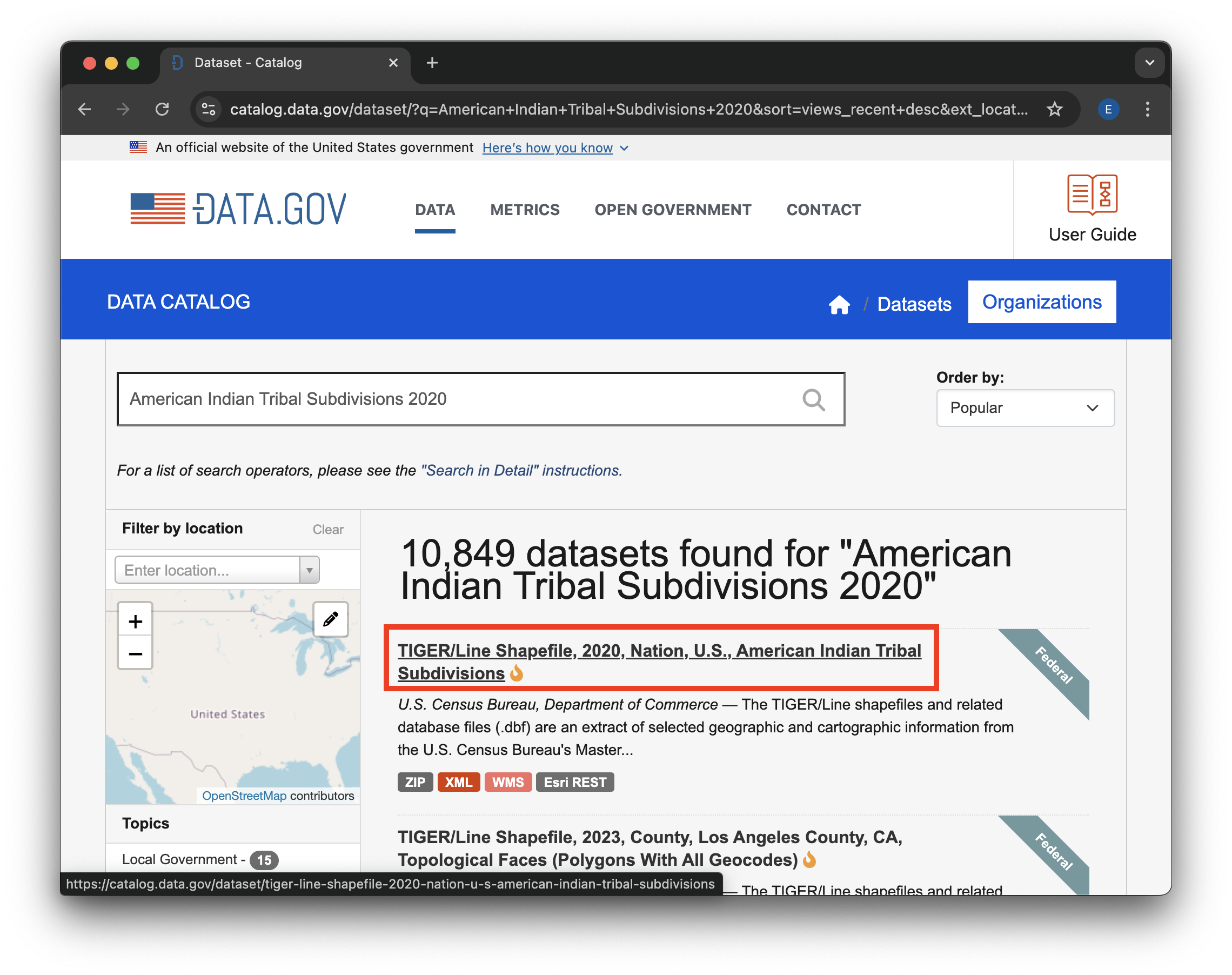
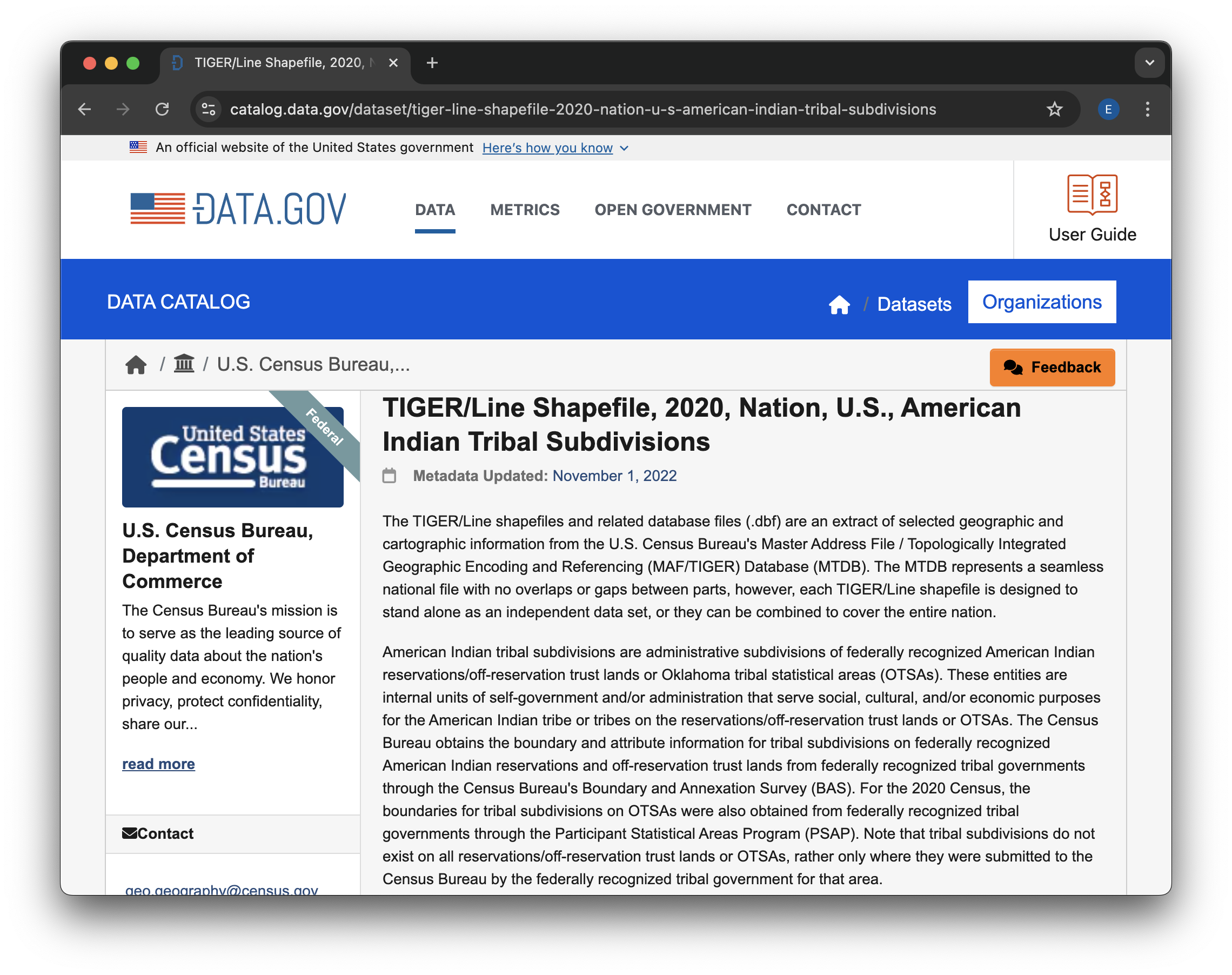
Find the “TIGER/Line Shapefile, 2020, Nation, U.S., American Indian Tribal Subdivisions” dataset in the search results, and click on the title to go to the dataset page.
 ## STEP 1D: Download
## STEP 1D: Download
Next, scroll down to the available files to download. Click on the Download button for the .zip file – this will be the easiest one to open in Python.
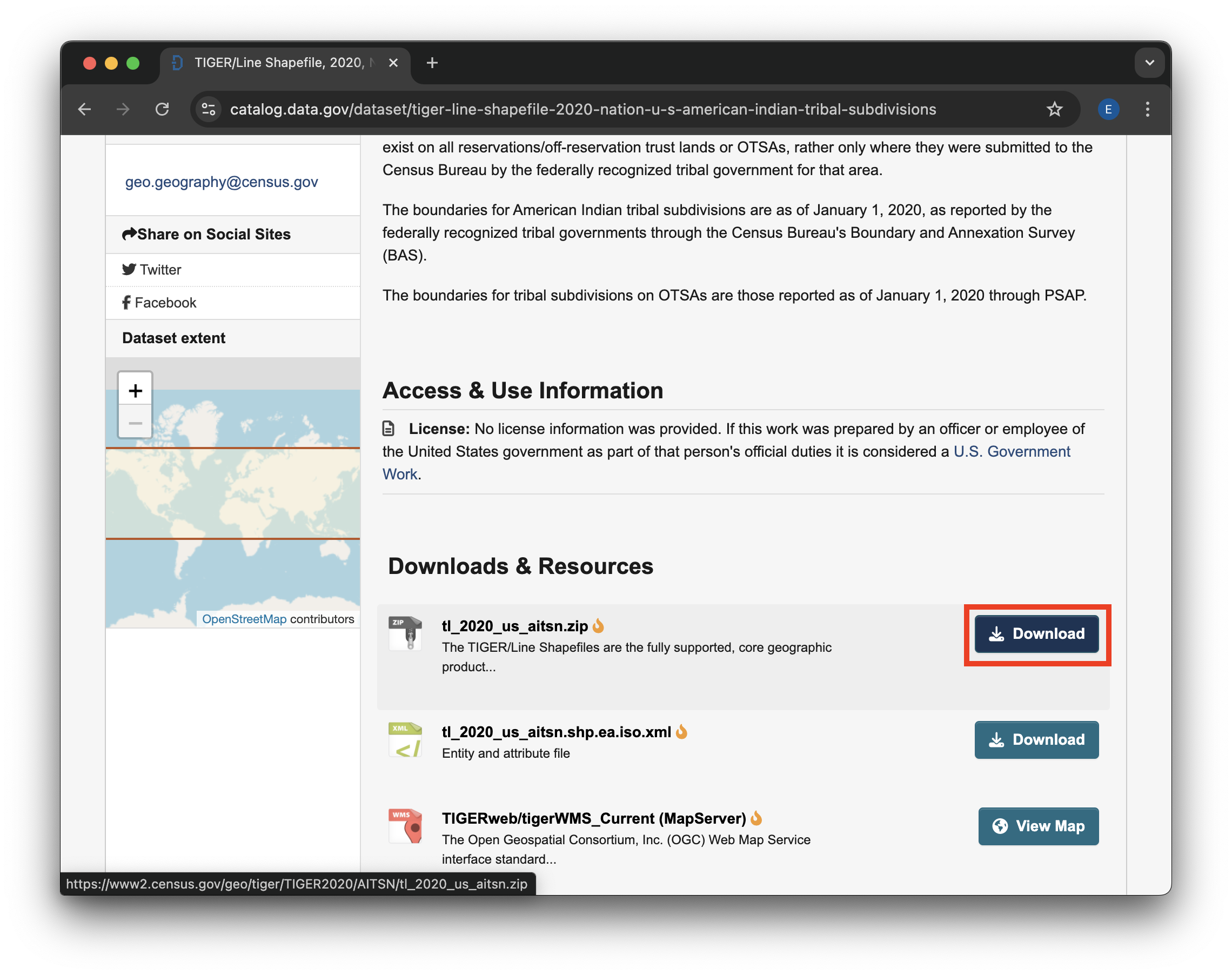
.zip file Download button.STEP 1E: Move your file
We now need to locate the file you downloaded and put it somewhere where Python can find it. Ideally, you should put the downloaded .zip file in your project directory. Most web browsers will pop up with some kind of button to open up your File Explorer (Windows) or Finder (Mac) in the location of your downloaded files. You can also check in your user home directory for a Downloads folder. If none of that works, try opening up your File Explorer/Finder and searching for the file
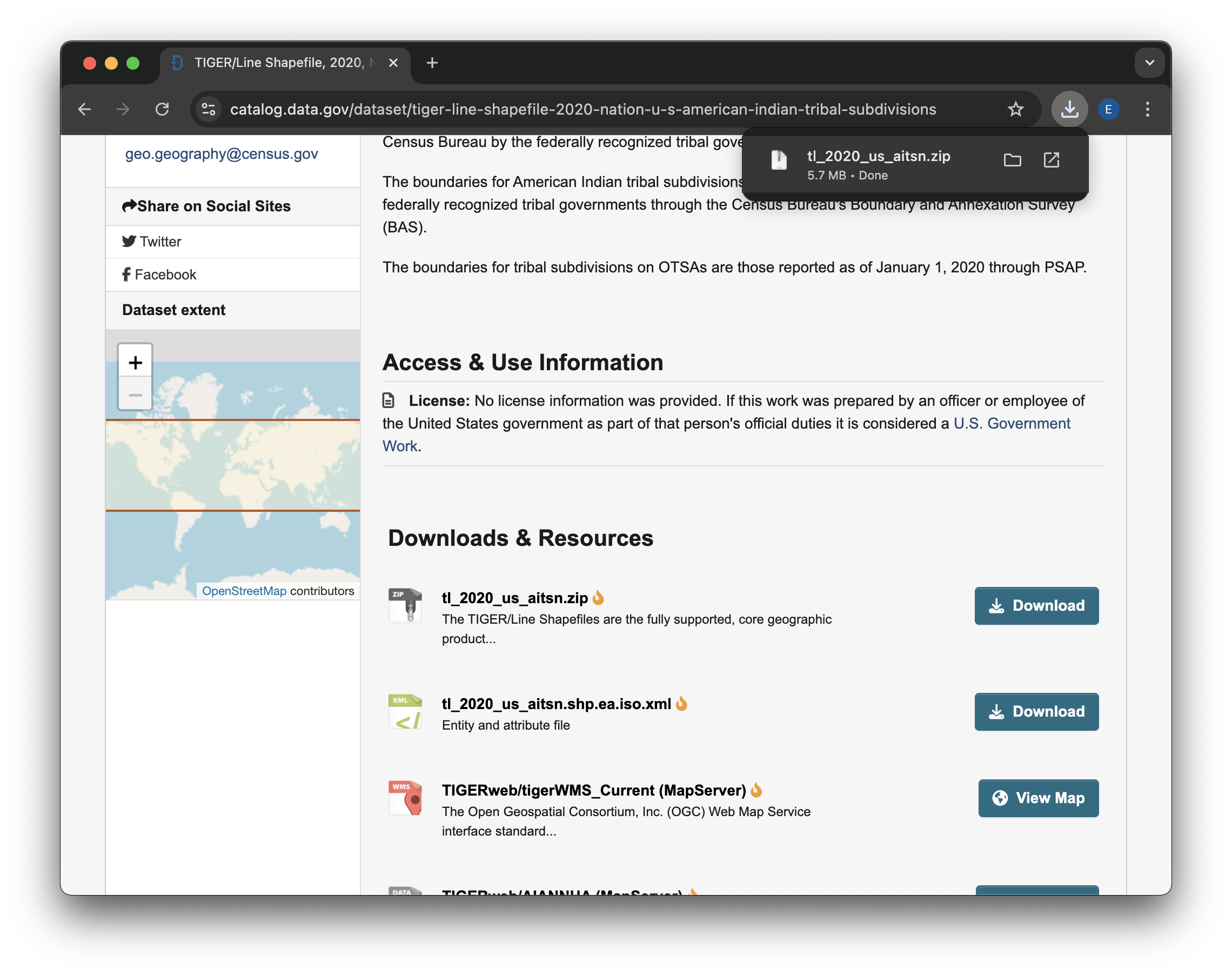
If you are working on GitHub Codespaces, you will need to upload your file before relocating it:
- Go to the
Explorertab in your Codespace - Right-click on the
datafolder - Click
Upload - Select the
.zipfile you just downloaded.
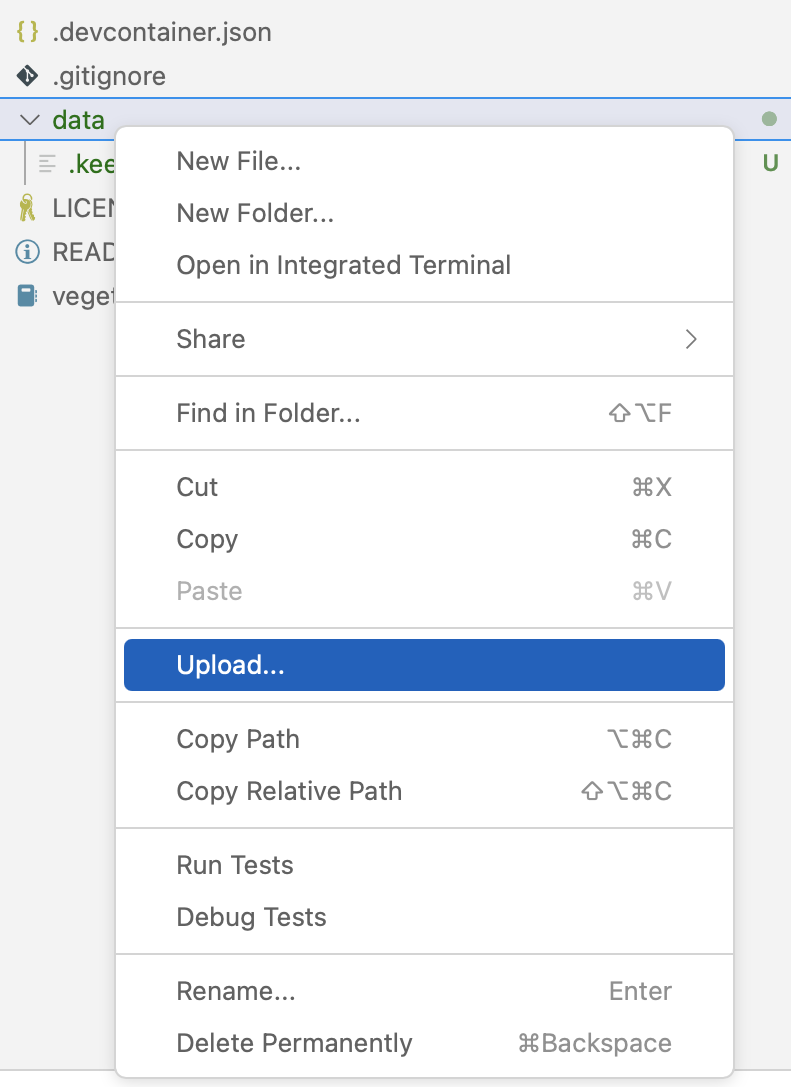
If you are working in GitHub Codespaces, you can skip ahead to unzipping the file! If not, you’ll need to relocate your file on your computer. One way to do that is with bash.
The code cell below is using a language called bash, which can be used to move files around your computer. To do that, we’re using the cp command, which stands for copy. In bash, you indicate that you want to retrieve a variable with the $ character. Since we’re in a Jupyter notebook, we can also access Python variables this way!
- Check that the path to your file,
~/Downloads/tl_2020_us_aitsn.zipis accurate, and change it if it isn’t - Run the cell to move your file.
!cp ~/Downloads/tl_2020_us_aitsn.zip -d "$project.project_dir"cp: cannot stat '/home/runner/Downloads/tl_2020_us_aitsn.zip': No such file or directoryNow, let’s check that the file got moved.
!ls "$project.project_dir"tl_2020_us_aitsn.zipYou can optionally unzip the file. geopandas will be able to read this particular shapefile from a .zip archive, but some other files may need to be unzipped. We’ll first define the path in Python, and then use bash to unzip. You can also unzip with the Python zipfile library, but it is more complicated than we need right now.
filename = "tl_2020_us_aitsn"
zip_path = project.project_dir / f"{filename}.zip"
unzip_dir = project.project_dir / filenameThe following command unzips a zip archive to the specified directory (-d). Any files that already exist are skipped without prompting (-n)
!unzip -n "$zip_path" -d "$unzip_dir"Archive: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn.zip
extracting: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn/tl_2020_us_aitsn.cpg
inflating: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn/tl_2020_us_aitsn.dbf
inflating: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn/tl_2020_us_aitsn.prj
inflating: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn/tl_2020_us_aitsn.shp
inflating: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn/tl_2020_us_aitsn.shp.ea.iso.xml
inflating: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn/tl_2020_us_aitsn.shp.iso.xml
inflating: /home/runner/.local/share/earth-analytics/gila-river/tl_2020_us_aitsn/tl_2020_us_aitsn.shx You can also delete the zip file, now that it is extracted. This will help keep your data directory tidy:
!rm "$zip_path"STEP 1F: Open boundary in Python
- Replace
"directory-name"with the actual name of the file you downloaded (or the folder you unzipped to) and put in your project directory. - Modify the code below to use descriptive variable names. Feel free to refer back to previous challenges for similar code!
- Add a line of code to open up the data path. What library and function do you need to open this type of data?
- Add some code to check your data, either by viewing it or making a quick plot. Does it look like what you expected?
# Define boundary path
path = project.project_dir / "directory-name"
# Open the site boundary
# Check that the data were downloaded correctlySee our solution!
# Define boundary path
aitsn_path = project.project_dir / "tl_2020_us_aitsn"
# Open the site boundary
aitsn_gdf = gpd.read_file(aitsn_path)
# Check that the data were downloaded correctly
aitsn_gdf.NAME0 Red Valley
1 Rock Point
2 Rough Rock
3 Indian Wells
4 Kayenta
...
479 1
480 Mission Highlands
481 Fort Thompson
482 Indian Point
483 Cheyenne and Arapaho District 2
Name: NAME, Length: 484, dtype: objectLet’s go ahead and select the Gila River subdivisions, and make a site map.
- Replace
identifierwith the value you found from exploring the interactive map. Make sure that you are using the correct data type! - Change the plot to have a web tile basemap, and look the way you want it to.
# Select and merge the subdivisions you want
# Plot the results with web tile imagesSee our solution!
# Select and merge the subdivisions you want
boundary_gdf = aitsn_gdf.loc[aitsn_gdf.AIANNHCE=='1310'].dissolve()
# Plot the results with web tile images
boundary_gdf.hvplot(
geo=True, tiles='EsriImagery',
fill_color=None, line_color='black',
title=site_name,
frame_width=500)STEP 2: AppEEARS API
Exploring the AppEEARS API for NASA Earthdata access
Before you get started with the data download today, you will need a free NASA Earthdata account if you don’t have one already!
Over the next four cells, you will download MODIS NDVI data for the study period. MODIS is a multispectral instrument that measures Red and NIR data (and so can be used for NDVI). There are two MODIS sensors on two different platforms: satellites Terra and Aqua.
Since we’re asking for a special download that only covers our study area, we can’t just find a link to the data - we have to negotiate with the data server. We’re doing this using the APPEEARS API (Application Programming Interface). The API makes it possible for you to request data using code. You can use code from the earthpy library to handle the API request.
Often when we want to do something more complex in coding we find an example and modify it. This download code is already almost a working example. Your task will be:
- Replace the start and end dates in the task parameters. Download data from July, when greenery is at its peak in the Northern Hemisphere.
- Replace the year range. You should get 3 years before and after the event so you can see the change!
- Replace
gdfwith the name of your site geodataframe. - Enter your NASA Earthdata username and password when prompted. The prompts can be a little hard to see – look at the top of your screen!
What would the product and layer name be if you were trying to download Landsat Surface Temperature Analysis Ready Data (ARD) instead of MODIS NDVI?
It can take some time for Appeears to process your request - anything from a few minutes to a few hours depending on how busy they are. You can check your progress by:
- Going to the Appeears webpage
- Clicking the
Exploretab - Logging in with your Earthdata account
# Initialize AppeearsDownloader for MODIS NDVI data
ndvi_downloader = eaapp.AppeearsDownloader(
download_key=download_key,
ea_dir=project.project_dir,
product='MOD13Q1.061',
layer='_250m_16_days_NDVI',
start_date="01-01",
end_date="01-31",
recurring=True,
year_range=[, ],
polygon=gdf
)
# Download the prepared download -- this can take some time!
ndvi_downloader.download_files(cache=True)See our solution!
# Initialize AppeearsDownloader for MODIS NDVI data
ndvi_downloader = eaapp.AppeearsDownloader(
download_key=download_key,
project=project,
product='MOD13Q1.061',
layer='_250m_16_days_NDVI',
start_date="06-01",
end_date="09-01",
recurring=True,
year_range=[start_year, end_year],
polygon=boundary_gdf
)
# Download the prepared download -- this can take some time!
ndvi_downloader.download_files(cache=True)Credentials found using 'env' backend.Putting it together: Working with multi-file raster datasets in Python
Now you need to load all the downloaded files into Python. You may have noticed that the `earthpy.appears module gives us all the downloaded file names…but only some of those are the NDVI files we want while others are quality files that tell us about the confidence in the dataset. For now, the files we want all have “NDVI” in the name.
Let’s start by getting all the NDVI file names. You will also need to extract the date from the filename. Check out the lesson on getting information from filenames in the textbook. We’re using a slightly different method here (the .rglob() or recursive glob method, which searchs all the directories nested inside the path), but the principle is the same.
glob doesn’t necessarily find files in the order you would expect. Make sure to sort your file names like it says in the textbook.
# Get a sorted list of NDVI tif file paths
ndvi_paths = sorted(list(project.project_dir.rglob('ndvi-pattern')))
# Display the first and last three files paths to check the pattern
ndvi_paths[:3], ndvi_paths[-3:]See our solution!
# Get a sorted list of NDVI tif file paths
ndvi_paths = sorted(list(project.project_dir.rglob('*NDVI*.tif')))
# Display the first and last three files paths to check the pattern
ndvi_paths[:3], ndvi_paths[-3:]([PosixPath('/home/runner/.local/share/earth-analytics/gila-river/gila-river-ndvi/MOD13Q1.061_2001137_to_2022244/MOD13Q1.061__250m_16_days_NDVI_doy2001145000000_aid0001.tif'),
PosixPath('/home/runner/.local/share/earth-analytics/gila-river/gila-river-ndvi/MOD13Q1.061_2001137_to_2022244/MOD13Q1.061__250m_16_days_NDVI_doy2001161000000_aid0001.tif'),
PosixPath('/home/runner/.local/share/earth-analytics/gila-river/gila-river-ndvi/MOD13Q1.061_2001137_to_2022244/MOD13Q1.061__250m_16_days_NDVI_doy2001177000000_aid0001.tif')],
[PosixPath('/home/runner/.local/share/earth-analytics/gila-river/gila-river-ndvi/MOD13Q1.061_2001137_to_2022244/MOD13Q1.061__250m_16_days_NDVI_doy2022209000000_aid0001.tif'),
PosixPath('/home/runner/.local/share/earth-analytics/gila-river/gila-river-ndvi/MOD13Q1.061_2001137_to_2022244/MOD13Q1.061__250m_16_days_NDVI_doy2022225000000_aid0001.tif'),
PosixPath('/home/runner/.local/share/earth-analytics/gila-river/gila-river-ndvi/MOD13Q1.061_2001137_to_2022244/MOD13Q1.061__250m_16_days_NDVI_doy2022241000000_aid0001.tif')])Repeating tasks in Python
Now you should have a few dozen files! For each file, you need to:
- Load the file in using the
rioxarraylibrary - Get the date from the file name
- Add the date as a dimension coordinate
- Give your data variable a name
You don’t want to write out the code for each file! That’s a recipe for copy pasta and errors. Luckily, Python has tools for doing similar tasks repeatedly. In this case, you’ll use one called a for loop.
There’s some code below that uses a for loop in what is called an accumulation pattern to process each file. That means that you will save the results of your processing to a list each time you process the files, and then merge all the arrays in the list.
- Look at the file names. How many characters from the end is the date?
doy_startanddoy_endare used to extract the day of the year (doy) from the file name. You will need to count characters from the end and change the values to get the right part of the file name. HINT: the index -1 in Python means the last value, -2 second-to-last, and so on. - Replace any required variable names with your chosen variable names
doy_start = -1
doy_end = -1
# Loop through each NDVI image
ndvi_das = []
for ndvi_path in ndvi_paths:
# Get date from file name
# Open dataset
# Add date dimension and clean up metadata
da = da.assign_coords({'date': date})
da = da.expand_dims({'date': 1})
da.name = 'NDVI'
# Prepare for concatenationSee our solution!
doy_start = -25
doy_end = -19
# Loop through each NDVI image
ndvi_das = []
for ndvi_path in ndvi_paths:
# Get date from file name
doy = ndvi_path.name[doy_start:doy_end]
date = pd.to_datetime(doy, format='%Y%j')
# Open dataset
da = rxr.open_rasterio(ndvi_path, mask_and_scale=True).squeeze()
# Add date dimension and clean up metadata
da = da.assign_coords({'date': date})
da = da.expand_dims({'date': 1})
da.name = 'NDVI'
# Prepare for concatenation
ndvi_das.append(da)
len(ndvi_das)154Next, stack your arrays by date into a time series:
- Modify the code to match your prior workflow steps and to use descriptive variable names
- Replace
coordinate_namewith the actual name of the coordinate you want to build up.
# Combine NDVI images from all dates
da = xr.combine_by_coords(list_of_data_arrays, coords=['coordinate_name'])
daSee our solution!
# Combine NDVI images from all dates
ndvi_da = xr.combine_by_coords(ndvi_das, coords=['date'])
ndvi_da<xarray.Dataset> Size: 48MB
Dimensions: (date: 154, y: 203, x: 382)
Coordinates:
band int64 8B 1
* x (x) float64 3kB -112.3 -112.3 -112.3 ... -111.5 -111.5 -111.5
* y (y) float64 2kB 33.39 33.39 33.38 33.38 ... 32.97 32.97 32.97
spatial_ref int64 8B 0
* date (date) datetime64[ns] 1kB 2001-01-14 2001-01-16 ... 2022-01-24
Data variables:
NDVI (date, y, x) float32 48MB 0.8282 0.6146 ... 0.2146 0.2085Your turn! Repeat this workflow in a different time and place.
It’s not only irrigation that affects NDVI! You could look at:
- Recovery after a national disaster, like a wildfire or hurricane
- Drought
- Deforestation
- Irrigation
- Beaver reintroduction
