id = 'stars'
site_name = 'Gila River Indian Community'
event = 'water rights case'
start_year = '2001'
end_year = '2022'
event_year = '2012'Water Rights Restored to the Gila River
The impacts of irrigation on vegetation health in the Gila River Valley
In 2004, the Akimel O’otham and Tohono O’odham tribes won a water rights settlement in the US Supreme Court. Using satellite imagery, we can see the effects of irrigation water on the local vegetation.
- Open raster or image data using code
- Combine raster data and vector data to crop images to an area of interest
- Summarize raster values with stastics
- Analyze a time-series of raster images

Reclaiming Water Rights on the Gila River
The Gila River Reservation south of Phoenix, AZ is the ancestral home of the Akimel O’otham and Tohono O’odham tribes. The Gila River area was known for its agriculture, with miles of canals providing irrigation. However, in the 1800s, European colonizers upstream installed dams which cut off water supply. This resulted in the collapse of Gila River agriculture, along with sky-rocketing rates of diabetes and heart disease in the community as they were force to subsist only on US government surplus rations.
In 2004, the Gila River community won back much of its water rights in court. The settlement granted senior water rights nearly matching pre-colonial water use. Work has begun to rebuild the agriculture in the Gila River Reservation. According to the Akimel O’otham and Tohono O’odham tribes, “It will take years to complete but in the end the community members will once again hear the sweet music of rushing water.”
Observing vegetation health from space
We will look at vegetation health using NDVI (Normalized Difference Vegetation Index). How does it work? First, we need to learn about spectral reflectance signatures.
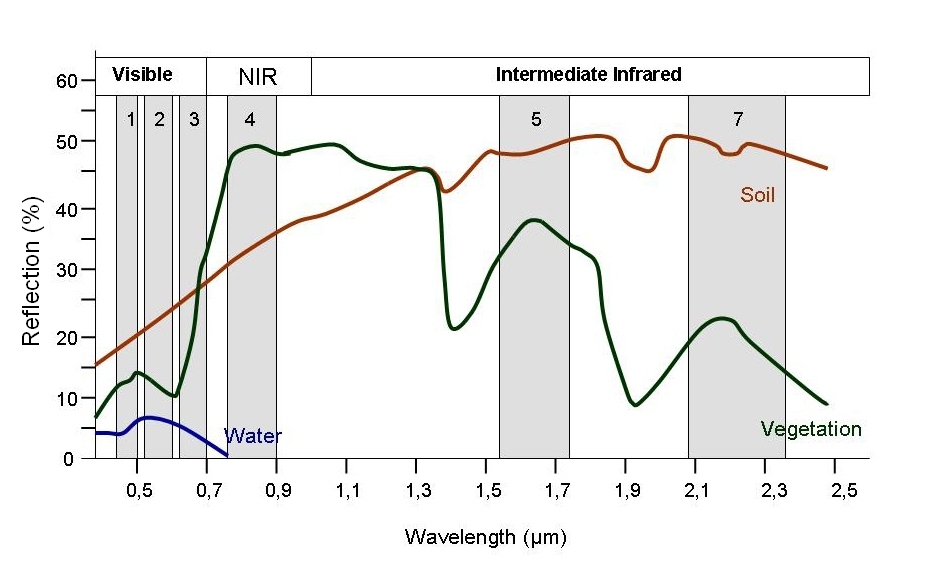
Every object reflects some wavelengths of light more or less than others. We can see this with our eyes, since, for example, plants reflect a lot of green in the summer, and then as that green diminishes in the fall they look more yellow or orange. The image below shows spectral signatures for water, soil, and vegetation:
 > Image source: SEOS Project
> Image source: SEOS Project
Healthy vegetation reflects a lot of Near-InfraRed (NIR) radiation. Less healthy vegetation reflects a similar amounts of the visible light spectra, but less NIR radiation. We don’t see a huge drop in Green radiation until the plant is very stressed or dead. That means that NIR allows us to get ahead of what we can see with our eyes.
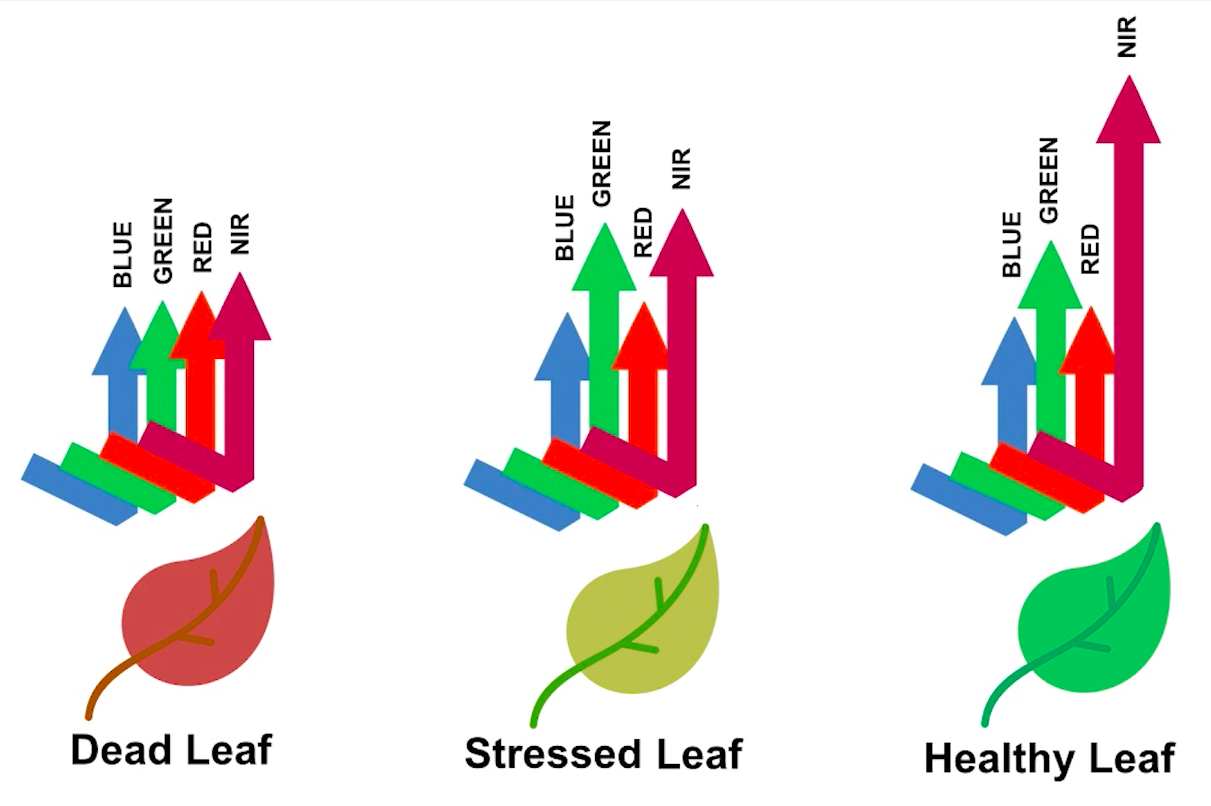 > Image source: Spectral signature literature review by px39n
> Image source: Spectral signature literature review by px39n
Different species of plants reflect different spectral signatures, but the pattern of the signatures across species and sitations is similar. NDVI compares the amount of NIR reflectance to the amount of Red reflectance, thus accounting for many of the species differences and isolating the health of the plant. The formula for calculating NDVI is:
\[NDVI = \frac{(NIR - Red)}{(NIR + Red)}\]
Read more about NDVI and other vegetation indices:
Before we get started coding, you can use the following parameters to change things about the workflow:
If the water rights case took place in 2004, why are we listing the event year as 2012? It takes time for practices to adjust. You can experiment with different years to see if it affects the results.
STEP 1: Site map
We’ll need some Python libraries to complete this workflow.
In the cell below, making sure to keep the packages in order, add packages for:
- Working with DataFrames
- Working with GeoDataFrames
- Making interactive plots of tabular and vector data
What are we using the rest of these packages for? See if you can figure it out as you complete the notebook.
import json
import os
import pathlib
import hvplot.xarray
import rioxarray as rxr
import xarray as xrSee our solution!
import json
from glob import glob
import earthpy
import geopandas as gpd
import hvplot.pandas
import hvplot.xarray
import pandas as pd
import rioxarray as rxr
import xarray as xrWe have one more setup task. We’re not going to be able to load all our data directly from the web to Python this time. That means we need to set up a place for it.
In the cell below, replace ‘my-data-folder’ with a descriptive directory name.
project = earthpy.Project("Gila River Vegetation", dirname='my-data-folder')
project.get_data()See our solution!
project = earthpy.Project("Gila River Vegetation")
project.get_data()
**Final Configuration Loaded:**
{}
🔄 Fetching metadata for article 29170118...
✅ Found 2 files for download.
[('https://ndownloader.figshare.com/files/54896600', 'tl_2020_us_aitsn', 'zip'), ('https://ndownloader.figshare.com/files/55242452', 'gila-river-ndvi', 'zip')]
tl_2020_us_aitsn
Downloading from https://ndownloader.figshare.com/files/54896600
Extracted output to /home/runner/.local/share/earth-analytics/gila-river-vegetation/tl_2020_us_aitsn
[PosixPath('/home/runner/.local/share/earth-analytics/gila-river-vegetation/tl_2020_us_aitsn')]
gila-river-ndvi
Downloading from https://ndownloader.figshare.com/files/55242452
Extracted output to /home/runner/.local/share/earth-analytics/gila-river-vegetation/gila-river-ndvi
[PosixPath('/home/runner/.local/share/earth-analytics/gila-river-vegetation/tl_2020_us_aitsn'), PosixPath('/home/runner/.local/share/earth-analytics/gila-river-vegetation/gila-river-ndvi')]Study Area: Gila River Indian Community
Earth Data Science data formats
In Earth Data Science, we get data in three main formats:
| Data type | Descriptions | Common file formats | Python type |
|---|---|---|---|
| Time Series | The same data points (e.g. streamflow) collected multiple times over time | Tabular formats (e.g. .csv, or .xlsx) | pandas DataFrame |
| Vector | Points, lines, and areas (with coordinates) | Shapefile (often an archive like a .zip file because a Shapefile is actually a collection of at least 3 files) |
geopandas GeoDataFrame |
| Raster | Evenly spaced spatial grid (with coordinates) | GeoTIFF (.tif), NetCDF (.nc), HDF (.hdf) |
rioxarray DataArray |
Check out the sections about about vector data and raster data in the textbook.
For this coding challenge, we are interested in the boundary of the Gila River Indian Community, and the health of vegetation in the area measured on a scale from -1 to 1. In the cell below, answer the following question: What data type do you think the boundary will be? What about the vegetation health?
Load the Gila River Indian Community boundary
- Locate the Tribal Subdivision files in your download directory
- Change
'subdivision-directory'to the actual location - Load the data into Python and check that it worked
# Load in the boundary data
aitsn_gdf = gpd.read_file(project.project_dir / 'subdivision-directory')
# Check that it workedSee our solution!
# Load in the boundary data
aitsn_gdf = gpd.read_file(project.project_dir / 'tl_2020_us_aitsn')
# Check that it worked
aitsn_gdf| AIANNHCE | TRSUBCE | TRSUBNS | GEOID | NAME | NAMELSAD | LSAD | CLASSFP | MTFCC | FUNCSTAT | ALAND | AWATER | INTPTLAT | INTPTLON | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2430 | 653 | 02419073 | 2430653 | Red Valley | Red Valley Chapter | T2 | D7 | G2300 | A | 922036695 | 195247 | +36.6294607 | -109.0550394 | POLYGON ((-109.2827 36.64644, -109.28181 36.65... |
| 1 | 2430 | 665 | 02419077 | 2430665 | Rock Point | Rock Point Chapter | T2 | D7 | G2300 | A | 720360268 | 88806 | +36.6598701 | -109.6166836 | POLYGON ((-109.85922 36.49859, -109.85521 36.5... |
| 2 | 2430 | 675 | 02419081 | 2430675 | Rough Rock | Rough Rock Chapter | T2 | D7 | G2300 | A | 364475668 | 216144 | +36.3976971 | -109.7695183 | POLYGON ((-109.93053 36.40672, -109.92923 36.4... |
| 3 | 2430 | 325 | 02418975 | 2430325 | Indian Wells | Indian Wells Chapter | T2 | D7 | G2300 | A | 717835323 | 133795 | +35.3248534 | -110.0855000 | POLYGON ((-110.24222 35.36327, -110.24215 35.3... |
| 4 | 2430 | 355 | 02418983 | 2430355 | Kayenta | Kayenta Chapter | T2 | D7 | G2300 | A | 1419241065 | 1982848 | +36.6884391 | -110.3045616 | POLYGON ((-110.56817 36.73489, -110.56603 36.7... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 479 | 1310 | 100 | 02418907 | 1310100 | 1 | District 1 | 28 | D7 | G2300 | N | 139902197 | 0 | +33.0600842 | -111.5806313 | POLYGON ((-111.63622 33.11798, -111.63405 33.1... |
| 480 | 4290 | 550 | 02612186 | 4290550 | Mission Highlands | Mission Highlands | 00 | D7 | G2300 | N | 6188043 | 0 | +48.0754384 | -122.2507432 | POLYGON ((-122.27579 48.07128, -122.27578 48.0... |
| 481 | 0855 | 400 | 02418941 | 0855400 | Fort Thompson | Fort Thompson District | 07 | D7 | G2300 | N | 535432708 | 38653364 | +44.1559680 | -099.4467700 | POLYGON ((-99.66452 44.25269, -99.66449 44.255... |
| 482 | 0335 | 300 | 02784108 | 0335300 | Indian Point | Indian Point Segment | T3 | D7 | G2300 | N | 326985 | 0 | +48.0604594 | -092.8466753 | POLYGON ((-92.85187 48.05944, -92.85186 48.059... |
| 483 | 5560 | 120 | 02804808 | 5560120 | Cheyenne and Arapaho District 2 | Cheyenne and Arapaho District 2 | 00 | D7 | G2300 | S | 4709525489 | 36177523 | +35.7613633 | -098.0107463 | POLYGON ((-98.61081 35.72524, -98.60732 35.725... |
484 rows × 15 columns
You might notice in this dataset that some of the names are not easily searchable. For example, the Gila River subdivisions are named “District 1-7”! So, how do we know what to search for? We recommend making an interactive plot of the data so that you can find the information you need, e.g.:
aitsn_gdf.hvplot(
geo=True, tiles='EsriImagery',
frame_width=500,
legend=False, fill_color=None, edge_color='white',
# This parameter makes all the column values in the dataset visible.
hover_cols='all')WARNING:param.main: edge_color option not found for polygons plot with bokeh; similar options include: ['muted_color', 'bgcolor', 'line_color']