import getpass
import json
import os
import pathlib
from glob import glob
import earthpy.appeears as eaapp
import hvplot.xarray
import rioxarray as rxr
import xarray as xrThe Cameron Peak Fire, Colorado, USA
The Cameron Peak Fire was the largest fire in Colorado history, with 326 square miles burned.
Observing vegetation health from space
We will look at the destruction and recovery of vegetation using NDVI (Normalized Difference Vegetation Index). How does it work? First, we need to learn about spectral reflectance signatures.
Every object reflects some wavelengths of light more or less than others. We can see this with our eyes, since, for example, plants reflect a lot of green in the summer, and then as that green diminishes in the fall they look more yellow or orange. The image below shows spectral signatures for water, soil, and vegetation:
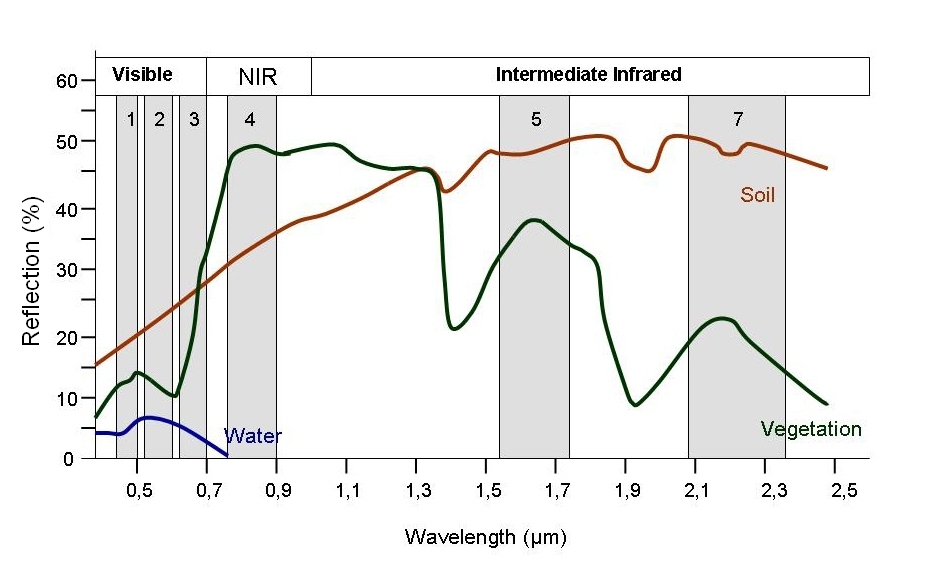 > Image source: SEOS Project
> Image source: SEOS Project
Healthy vegetation reflects a lot of Near-InfraRed (NIR) radiation. Less healthy vegetation reflects a similar amounts of the visible light spectra, but less NIR radiation. We don’t see a huge drop in Green radiation until the plant is very stressed or dead. That means that NIR allows us to get ahead of what we can see with our eyes.
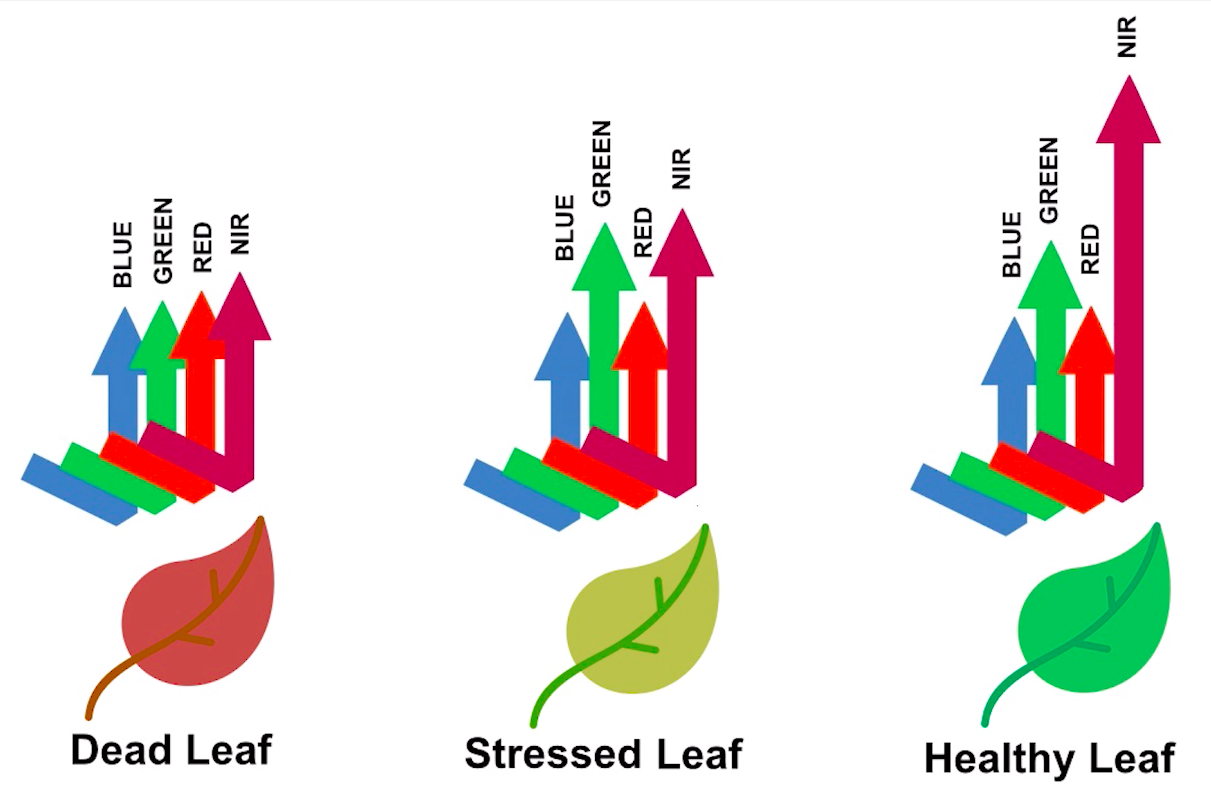 > Image source: Spectral signature literature review by px39n
> Image source: Spectral signature literature review by px39n
Different species of plants reflect different spectral signatures, but the pattern of the signatures are similar. NDVI compares the amount of NIR reflectance to the amount of Red reflectance, thus accounting for many of the species differences and isolating the health of the plant. The formula for calculating NDVI is:
\[NDVI = \frac{(NIR - Red)}{(NIR + Red)}\]
Read more about NDVI and other vegetation indices:
See our solution!
import getpass
import json
import os
import pathlib
from glob import glob
import earthpy.appeears as eaapp
import geopandas as gpd
import hvplot.pandas
import hvplot.xarray
import pandas as pd
import rioxarray as rxr
import xarray as xrWe have one more setup task. We’re not going to be able to load all our data directly from the web to Python this time. That means we need to set up a place for it.
data_dir = os.path.join(pathlib.Path.home(), 'my-data-folder')
# Make the data directory
os.makedirs(data_dir, exist_ok=True)See our solution!
data_dir = os.path.join(
pathlib.Path.home(), 'earth-analytics', 'data', 'cameron-peak')
# Make the data directory
os.makedirs(data_dir, exist_ok=True)Study Area: Cameron Peak Fire Boundary
Earth Data Science data formats
In Earth Data Science, we get data in three main formats:
| Data type | Descriptions | Common file formats | Python type |
|---|---|---|---|
| Time Series | The same data points (e.g. streamflow) collected multiple times over time | Tabular formats (e.g. .csv, or .xlsx) | pandas DataFrame |
| Vector | Points, lines, and areas (with coordinates) | Shapefile (often an archive like a .zip file because a Shapefile is actually a collection of at least 3 files) |
geopandas GeoDataFrame |
| Raster | Evenly spaced spatial grid (with coordinates) | GeoTIFF (.tif), NetCDF (.nc), HDF (.hdf) |
rioxarray DataArray |
# Download the Cameron Peak fire boundarySee our solution!
url = (
"https://services3.arcgis.com/T4QMspbfLg3qTGWY/arcgis/rest/services"
"/WFIGS_Interagency_Perimeters/FeatureServer/0/query"
"?where=poly_IncidentName%20%3D%20'CAMERON%20PEAK'"
"&outFields=*&outSR=4326&f=json")
gdf = gpd.read_file(url)
gdf| OBJECTID | poly_SourceOID | poly_IncidentName | poly_FeatureCategory | poly_MapMethod | poly_GISAcres | poly_CreateDate | poly_DateCurrent | poly_PolygonDateTime | poly_IRWINID | ... | attr_Source | attr_IsCpxChild | attr_CpxName | attr_CpxID | attr_SourceGlobalID | GlobalID | Shape__Area | Shape__Length | attr_IncidentComplexityLevel | geometry | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 14659 | 10393 | Cameron Peak | Wildfire Final Fire Perimeter | Infrared Image | 208913.3 | NaT | 2023-03-14 15:13:42.810000+00:00 | 2020-10-06 21:30:00+00:00 | {53741A13-D269-4CD5-AF91-02E094B944DA} | ... | FODR | None | None | None | {5F5EC9BA-5F85-495B-8B97-1E0969A1434E} | e8e4ffd4-db84-4d47-bd32-d4b0a8381fff | 0.089957 | 5.541765 | None | MULTIPOLYGON (((-105.88333 40.54598, -105.8837... |
1 rows × 115 columns
ans_gdf = _
gdf_pts = 0
if isinstance(ans_gdf, gpd.GeoDataFrame):
print('\u2705 Great work! You downloaded and opened a GeoDataFrame')
gdf_pts +=2
else:
print('\u274C Hmm, your answer is not a GeoDataFrame')
print('\u27A1 You earned {} of 2 points for downloading data'.format(gdf_pts))✅ Great work! You downloaded and opened a GeoDataFrame
➡ You earned 2 of 2 points for downloading dataSite Map
We always want to create a site map when working with geospatial data. This helps us see that we’re looking at the right location, and learn something about the context of the analysis.
# Plot the Cameron Peak Fire boundarygdf.hvplot(
geo=True,
title='Cameron Peak Fire, 2020',
tiles='EsriImagery')Exploring the AppEEARS API for NASA Earthdata access
Before you get started with the data download today, you will need a free NASA Earthdata account if you don’t have one already!
Over the next four cells, you will download MODIS NDVI data for the study period. MODIS is a multispectral instrument that measures Red and NIR data (and so can be used for NDVI). There are two MODIS sensors on two different platforms: satellites Terra and Aqua.
Since we’re asking for a special download that only covers our study area, we can’t just find a link to the data - we have to negotiate with the data server. We’re doing this using the APPEEARS API (Application Programming Interface). The API makes it possible for you to request data using code. You can use code from the earthpy library to handle the API request.
It can take some time for Appeears to process your request - anything from a few minutes to a few hours depending on how busy they are. You can check your progress by:
- Going to the Appeears webpage
- Clicking the
Exploretab - Logging in with your Earthdata account
# Initialize AppeearsDownloader for MODIS NDVI data
ndvi_downloader = eaapp.AppeearsDownloader(
download_key='cp-ndvi',
ea_dir=data_dir,
product='MOD13Q1.061',
layer='_250m_16_days_NDVI',
start_date="01-01",
end_date="01-31",
recurring=True,
year_range=[2021, 2021],
polygon=gdf
)
# Download the prepared download -- this can take some time!
ndvi_downloader.download_files(cache=True)# Initialize AppeearsDownloader for MODIS NDVI data
ndvi_downloader = eaapp.AppeearsDownloader(
download_key='cp-ndvi',
ea_dir=data_dir,
product='MOD13Q1.061',
layer='_250m_16_days_NDVI',
start_date="07-01",
end_date="07-31",
recurring=True,
year_range=[2018, 2023],
polygon=gdf
)
ndvi_downloader.download_files(cache=True)** Message: 18:00:50.035: Remote error from secret service: org.freedesktop.DBus.Error.ServiceUnknown: The name org.freedesktop.secrets was not provided by any .service filesPutting it together: Working with multi-file raster datasets in Python
Now you need to load all the downloaded files into Python. Let’s start by getting all the file names. You will also need to extract the date from the filename. Check out the lesson on getting information from filenames in the textbook.
# Get a list of NDVI tif file pathsSee our solution!
# Get list of NDVI tif file paths
ndvi_paths = sorted(glob(os.path.join(data_dir, 'cp-ndvi', '*', '*NDVI*.tif')))
len(ndvi_paths)18Repeating tasks in Python
Now you should have a few dozen files! For each file, you need to:
- Load the file in using the
rioxarraylibrary - Get the date from the file name
- Add the date as a dimension coordinate
- Give your data variable a name
- Divide by the scale factor of 10000
You don’t want to write out the code for each file! That’s a recipe for copy pasta. Luckily, Python has tools for doing similar tasks repeatedly. In this case, you’ll use one called a for loop.
There’s some code below that uses a for loop in what is called an accumulation pattern to process each file. That means that you will save the results of your processing to a list each time you process the files, and then merge all the arrays in the list.
scale_factor = 1
doy_start = -1
doy_end = -1See our solution!
scale_factor = 10000
doy_start = -19
doy_end = -12ndvi_das = []
for ndvi_path in ndvi_paths:
# Get date from file name
doy = ndvi_path[doy_start:doy_end]
date = pd.to_datetime(doy, format='%Y%j')
# Open dataset
da = rxr.open_rasterio(ndvi_path, masked=True).squeeze()
# Add date dimension and clean up metadata
da = da.assign_coords({'date': date})
da = da.expand_dims({'date': 1})
da.name = 'NDVI'
# Multiple by scale factor
da = da / scale_factor
# Prepare for concatenation
ndvi_das.append(da)
len(ndvi_das)18Next, stack your arrays by date into a time series using the xr.combine_by_coords() function. You will have to tell it which dimension you want to stack your data in.
See our solution!
ndvi_da = xr.combine_by_coords(ndvi_das, coords=['date'])
ndvi_da<xarray.Dataset> Size: 4MB
Dimensions: (date: 18, y: 153, x: 339)
Coordinates:
band int64 8B 1
* x (x) float64 3kB -105.9 -105.9 -105.9 ... -105.2 -105.2 -105.2
* y (y) float64 1kB 40.78 40.78 40.77 40.77 ... 40.47 40.46 40.46
spatial_ref int64 8B 0
* date (date) datetime64[ns] 144B 2018-06-26 2018-07-12 ... 2023-07-28
Data variables:
NDVI (date, y, x) float32 4MB 0.5672 0.5711 0.6027 ... 0.6327 0.6327# Compute the difference in NDVI before and after the fire
# Plot the difference
(
ndvi_diff.hvplot(x='', y='', cmap='', geo=True)
*
gdf.hvplot(geo=True, fill_color=None, line_color='black')
)--------------------------------------------------------------------------- NameError Traceback (most recent call last) Cell In[17], line 5 1 # Compute the difference in NDVI before and after the fire 2 3 # Plot the difference 4 ( ----> 5 ndvi_diff.hvplot(x='', y='', cmap='', geo=True) 6 * 7 gdf.hvplot(geo=True, fill_color=None, line_color='black') 8 ) NameError: name 'ndvi_diff' is not defined
See our solution!
ndvi_diff = (
ndvi_da
.sel(date=slice('2021', '2023'))
.mean('date')
.NDVI
- ndvi_da
.sel(date=slice('2018', '2020'))
.mean('date')
.NDVI
)
(
ndvi_diff.hvplot(x='x', y='y', cmap='PiYG', geo=True)
*
gdf.hvplot(geo=True, fill_color=None, line_color='black')
)Is the NDVI lower within the fire boundary after the fire?
You will compute the mean NDVI inside and outside the fire boundary. First, use the code below to get a GeoDataFrame of the area outside the Reservation. Your task: * Check the variable names - Make sure that the code uses your boundary GeoDataFrame * How could you test if the geometry was modified correctly? Add some code to take a look at the results.
out_gdf = (
gpd.GeoDataFrame(geometry=gdf.envelope)
.overlay(gdf, how='difference'))Next, clip your DataArray to the boundaries for both inside and outside the reservation. You will need to replace the GeoDataFrame name with your own. Check out the lesson on clipping data with the rioxarray library in the textbook.
# Clip data to both inside and outside the boundarySee our solution!
ndvi_cp_da = ndvi_da.rio.clip(gdf.geometry, from_disk=True)
ndvi_out_da = ndvi_da.rio.clip(out_gdf.geometry, from_disk=True)Finally, plot annual July means for both inside and outside the Reservation on the same plot.
:::
# Compute mean annual July NDVISee our solution!
# Compute mean annual July NDVI
jul_ndvi_cp_df = (
ndvi_cp_da
.groupby(ndvi_cp_da.date.dt.year)
.mean(...)
.NDVI.to_dataframe())
jul_ndvi_out_df = (
ndvi_out_da
.groupby(ndvi_out_da.date.dt.year)
.mean(...)
.NDVI.to_dataframe())
# Plot inside and outside the reservation
jul_ndvi_df = (
jul_ndvi_cp_df[['NDVI']]
.join(
jul_ndvi_out_df[['NDVI']],
lsuffix=' Burned Area', rsuffix=' Unburned Area')
)
jul_ndvi_df.hvplot(
title='NDVI before and after the Cameron Peak Fire'
)Now, take the difference between outside and inside the Reservation and plot that. What do you observe? Don’t forget to write a headline and description of your plot!
# Plot difference inside and outside the reservationSee our solution!
# Plot difference inside and outside the reservation
jul_ndvi_df['difference'] = (
jul_ndvi_df['NDVI Burned Area']
- jul_ndvi_df['NDVI Unburned Area'])
jul_ndvi_df.difference.hvplot(
title='Difference between NDVI within and outside the Cameron Peak Fire'
)Your turn! Repeat this workflow in a different time and place.
It’s not just fires that affect NDVI! You could look at:
- Recovery after a national disaster, like a wildfire or hurricane
- Drought
- Deforestation
- Irrigation
- Beaver reintroduction